Publications
For a full list see Google Scholar

Iasi et al.
In this joint paper with the Moorjani lab, we provide a systematic overview how Neandertal ancestry in early modern humans changed through time. We estimate that the gene flow happened around 47,000 years ago, and lasted around 7,000 years. We find that all Neandertal ancestry in people living today traces back to this event, and that selection-both positive and negative-on Neandertal ancestry happened very rapidly.

Sümer et al.
In this paper, led by Arev, Kay Prüfer and Johannes Krause, we provide new genetic data from early modern humans from Zlaty Kun, Czechia and Ranis, Germany. We show that they were members of a small population that went extinct, and that the individuals at Ranis and Zlaty Kun were close relatives.

D Popli, BM Peter
In this paper, we develop a statistical framework to study population structure. We show that different versions of PCA yield different interpretations in terms of noise vs. population structure. Our results suggest that probablistic versions of PCA are better suited to revealing population structure, compared to the methods currently employed.
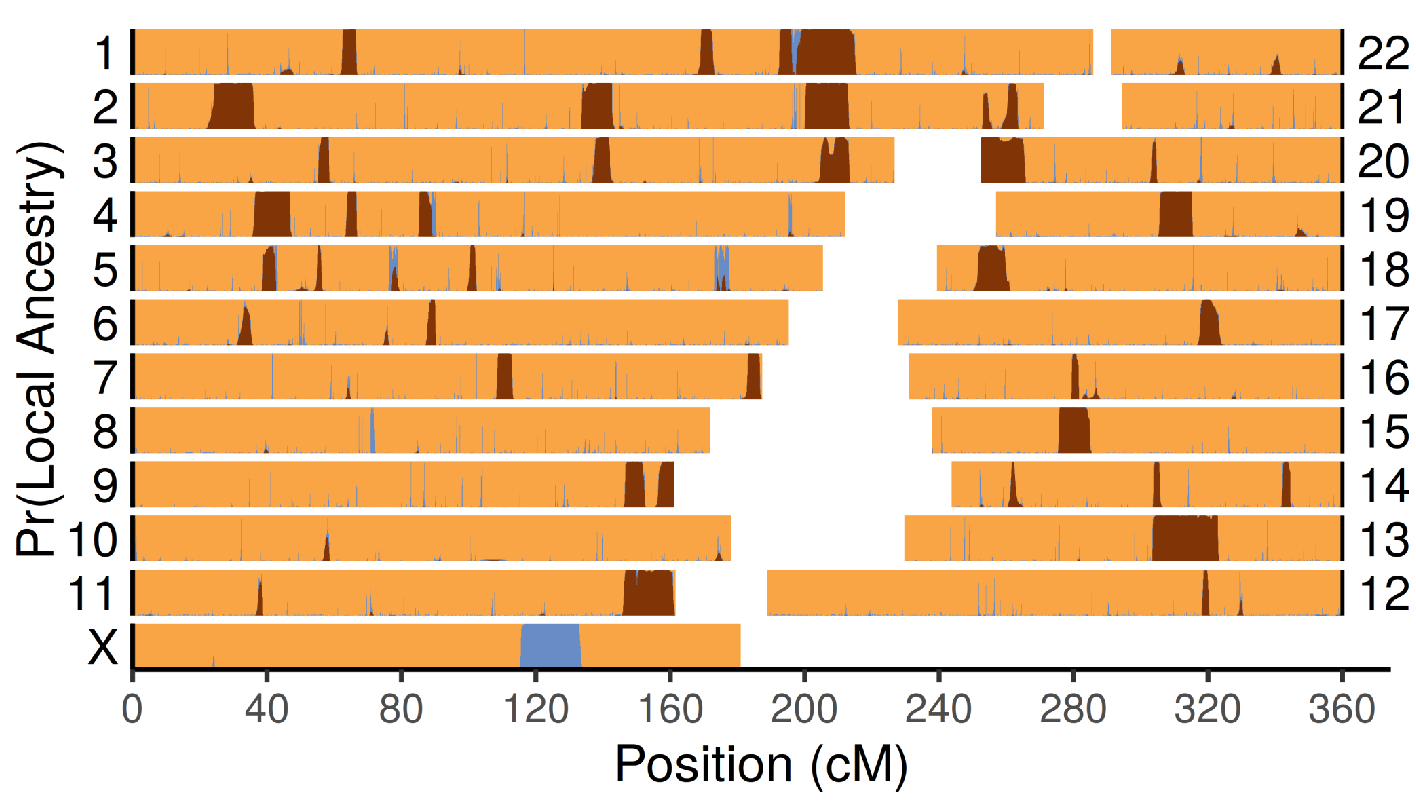
BM Peter
I developed a new method to detect local ancestry in archaic genomes
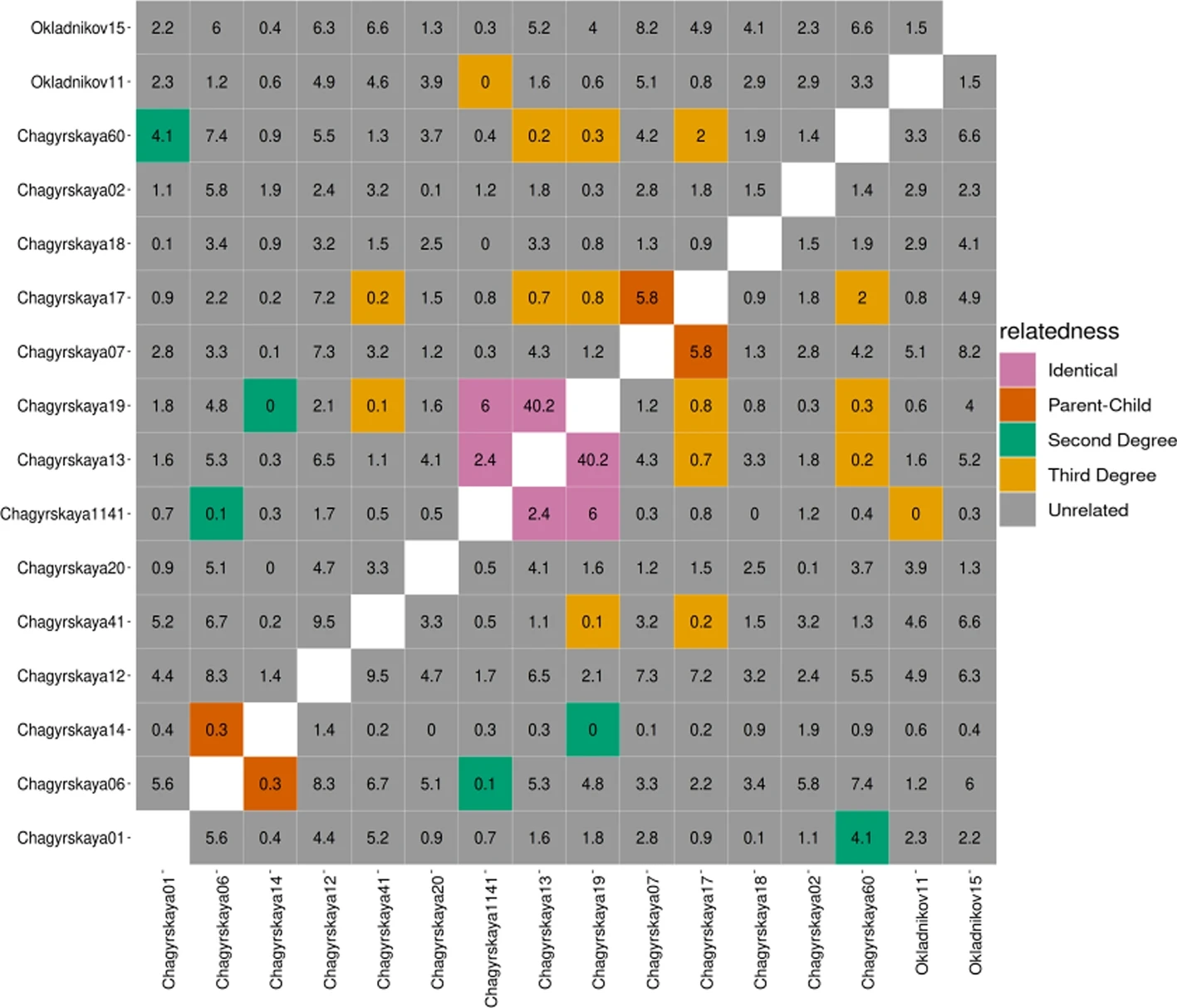
Divyaratan Popli, Stéphane Peyrégne, BM Peter
We develop a new method to classify familial relatedness up to 3rd degree using minimal data, taking contamination and ancient DNA damage into account

Skov et al.
We develop a new method to classify familial relatedness up to 3rd degree using minimal data, taking contamination and ancient DNA damage into account
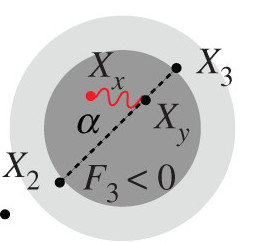
Peter BM
Philos. Trans. R. Soc. B (2022)
I develop some theory that show that F-statistics and PCA belong to the same framework. This allows us to interpret PCA-plots quantitatively in the context of admixture
Iasi, LNM, Ringbauer H & Peter BM
Molecular Biology and Evolution (2021)
Over which time period did Humans and Neandertals interact? In this paper, we develop theory to show that this question is unanswerable from present-day genetic data

Massilani D, Skov L, Hajdinjak M, Gunchinsuren B, Tseveendorj D, Yi S, Lee J, Nagel S, Nickel B, Devies T, Higham T, Meyer M, Kelso J, Peter BM, Paabo S
We contribute to a paper describing the oldest Human from Mongolia, who lived more than 34,000 years ago. We show that he already had Denisovan ancestry, and that the same ancestry persists in present-day East Asians.
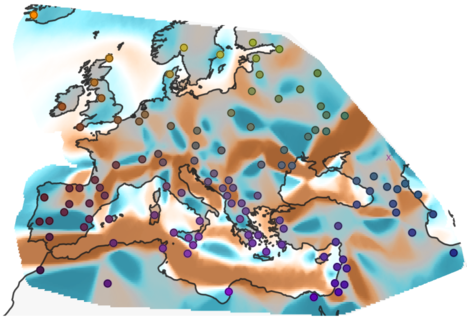
Peter, BM, Petkova D , Novembre J
Molecular Biology and Evolution (2020)
We show that Human Genetic Diversity can (largely) be explained by spatial constraints; and that these constraints are commonly mountains, deserts, oceans, i.e. barriers to gene flow and migration.
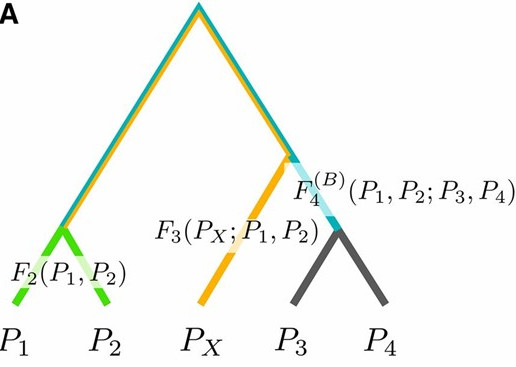
Peter BM
I develop and explain the population genetic background of F-statistics, a popular and widely used framework to analyze complex population structure.
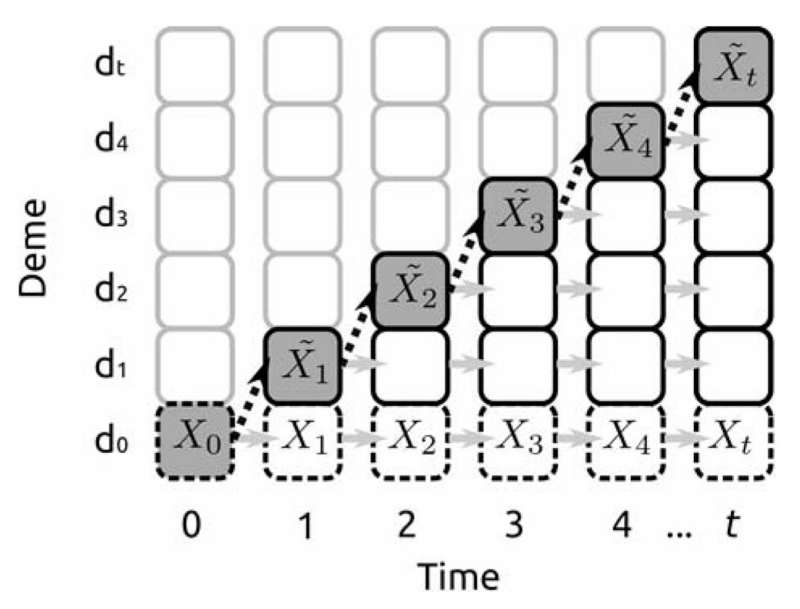
Peter BM and Slatkin M
I describe a simple model to define an effective founder size, which measures how much a population’s diversity declines during an expansion
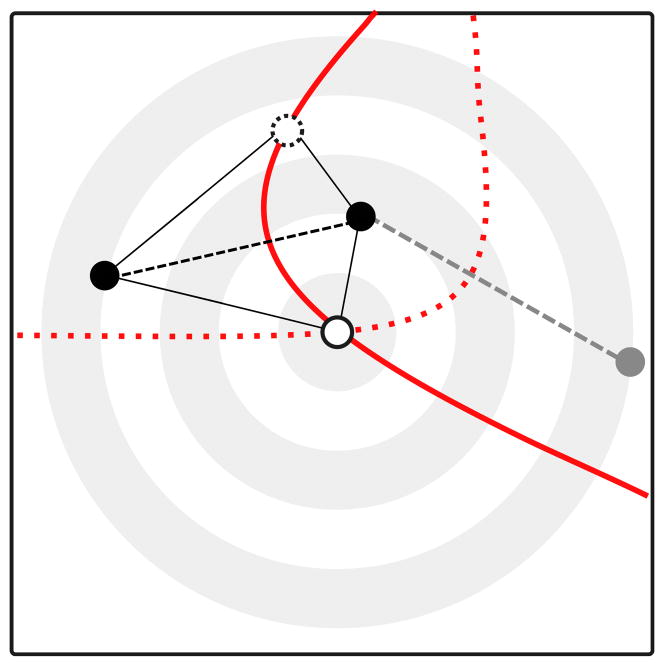
Peter BM and Slatkin M
We develop a statistic, called the directionality index, that allows the characterization of the direction of gene flow from genetic data. It also allows pinpointing the origin of an expansion.
Full List of publications
A joint framework for studying population structure using principal component analysis and F-statistics (2024)
Leonardo N. M. Iasi1, Manjusha Chintalapati, Laurits Skov, Alba Bossoms Mesa, Mateja Hajdinjak, Benjamin M. Peter, Priya Moorjani
Science
Earliest modern human genomes constrain timing of Neanderthal admixture (2024)
Sümer et al.
Nature
A joint framework for studying population structure using principal component analysis and F-statistics (2024)
Divyaratan Popli, BM Peter
biorxiv
100,000 years of gene flow between Neandertals and Denisovans in the Altai mountains. ()
BM Peter
biorxiv
KIN: A method to infer relatedness from low-coverage ancient DNA (2023)
Divyaratan Popli, Stéphane Peyrégne, BM Peter
Genome Biology
Genetic insights into the social organization of Neanderthals (2022)
Skov L, Peyrégne S, Popli D, Iasi LNM, Devièse T, Slon V, Zavala EI, Hajdinjak M, Sümer AP, Grote S, Bossoms Mesa A, López Herráez D, Nickel B, Nagel S, Richter J, Essel E, Gansauge M, Schmidt A, Korlević P, Comeskey D, Derevianko AP, Kharevich A, Markin SV, Talamo S, Douka K, Krajcarz MT, Roberts RG, Higham T, Viola B, Krivoshapkin AI, Kolobova KA, Kelso J, Meyer M, Pääbo S Peter, BM.
Nature
Assessing human genome-wide variation in the Massim region of Papua New Guinea and implications for the Kula trading tradition. (2022)
*Liu D, Peter BM, Schiefenhovel W, Kayser M, Stoneking M
Molecular Biology and Evolution
The evolutionary history of human spindle genes includes back-and-forth gene flow with Neandertals (2022)
Peyrégne S, Kelso J, Peter BM, Pääbo S
eLife
A geometric relationship of F2, F3 and F4-statistics with principal component analysis. (2022)
Peter BM
Philos. Trans. R. Soc. B
An extended admixture pulse model reveals the limitations to Human-Neandertal introgression dating (2021)
Iasi, LNM, Ringbauer H & Peter BM
Molecular Biology and Evolution
Mitonuclear interactions and introgression genomics of macaque monkeys [Macaca] highlight the influence of behaviour on genome evolution (2021)
Evans BJ, Peter BM, Melnick DJ, Andayani N, Supriatna J, Zhu J & Tosi AJ
Proc. Roy. Soc. B
Different historical generation intervals in human populations inferred from Neanderthal fragment lengths and patterns of mutation accumulation. (2021)
Coll Macià M, Skov L, Peter, BM, Schierup MH
Nature Communications
Genome of a middle Holocene hunter-gatherer from Wallacea. (2021)
Carlhoff S, Duli A, Nägele K, Nur M, Skov L, Sumantri I, Oktaviana AA, Hakim B, Burhan B, Syahdar FA, McGahan DP, Bulbeck D, Perston YL, Newman K, Saiful AM, Ririmasse M, Chia S, Hasanuddin, Suryatman DATP, Supriaadi, Jeong C, Peter BM, Prüfer K, Powell A, Krause J, Posth C, Brumm A
Nature
Initial Upper Palaeolithic humans in Europe had recent Neanderthal ancestry. (2021)
Hajdinjak M, Mafessoni F, Skov L, Vernot B, Hubner A, Fu Q, Essel E, Nagel S, Nickel B, Richter J, Moldovan OT, Constation S,, Endarova E, Zahariev N, Spasov R, Welker F, Smitt GM, Sinet-Mathiot V, Paskulin L, Fewlass H, Talamo S, Rezek Z, Sirakova S, Sirakov N, McPherron SP, Tsanova T, Hublin JJ, Peter BM, Meyer M, Skoglund P, Kelso J, Paabo S
Nature
Unearthing Neanderthal population history using nuclear and mitochondrial DNA from cave sediments. (2021)
Vernot B, Zavala EI, Gómez-Olivencia A, Jacobs Z, Slon V, Mafessoni F, Romagné F, Pearson A, Petr M, Sala N, Pablos A, Aranburu A, Bermúdez de Castro JM, Carbonell E, Li B, Krajcarz MT, Krivoshapkin AI, Kolobova KA, Kozlikin MB, Shunkov MV, Derevianko AP, Viola B, Grote S, Essel E, López Herráez D, Nagel S, Nickel B, Richter J, Schmidt A, Peter BM, Kelso J, Roberts RG, Arsuaga JL, Meyer M
Science
Denisovan ancestry and population history of early East Asians (2020)
Massilani D, Skov L, Hajdinjak M, Gunchinsuren B, Tseveendorj D, Yi S, Lee J, Nagel S, Nickel B, Devies T, Higham T, Meyer M, Kelso J, Peter BM, Paabo S
Science
AuthentiCT: a model of ancient DNA damage to estimate the proportion of present-day DNA contamination. (2020)
Peyrėgne S & Peter BM
Genome Biology
A high-coverage Neandertal genome from Chagyrskaya Cave (2020)
Mafessoni F, Grote S, de Filippo C, Slon V, Kolobova K, Viola, B, Markin S, Chintalapati M, Peyrégne S, Skov L, Skoglund P, Krivoshapkin A Derevianko A, Meyer M, Kelso B, Peter B, Prüfer K & Pääbo S
PNAS
Genetic landscapes reveal how human genetic diversity aligns with geography (2020)
Peter, BM, Petkova D , Novembre J
Molecular Biology and Evolution
Human local adaptation of the TRPM8 cold receptor along a latitudinal cline (2018)
Key FM, Muslihudeen AA, Mundry R, Peter BM, D’Amato M, Dennis MY, Schmidt JM and Andrés, AM
PLoS Genetics
A longitudinal cline characterizes the genetic structure of human populations in the Tibetan plateau. (2017)
Jeong C, Peter BM, Basnyat B, Neupane M, Beall CM, Childs G, Craig SR, Novembre J and Di Rienzo A
PLoS One
Chimpanzee genomic diversity reveals ancient admixture with bonobos (2016)
Marc de Manuel M, Kuhlwilm M, Frandsen P, et al. [and 34 others, including Peter BM]
Science
Recent advances in the study of fine-scale population structure in humans. (2016)
Novembre J, and **Peter BM
Curr. Opin. Genet. Dev.
Admixture, Population Structure, and F-Statistics (2016)
Peter BM
Genetics
Phylogenomics at the tips: inferring lineages and their demographic history in a tropical lizard, Carlia amax (2016)
Potter S, Bragg J, Peter, BM, Bi K, Moritz C
Molecular Ecology
Estimating the ages of selection signals from different epochs in human history (2015)
Nakagome S, Alkorta-Aranburu G, Amato R, Howie B, Peter BM, Hudson RR and Di Rienzo A
Molecular Biology and Evolution
The effective founder effect in a spatially expanding population (2015)
Peter BM and Slatkin M
Evolution
Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA (2014)
Huerta-Sánchez E, Jin X, Bianba Z, Peter BM et al.
Nature
Selection on a Variant Associated with Improved Viral Clearance Drives Local Adaptive Pseudogenization of Interferon Lambda 4 (IFNL4). (2014)
Key FM, Peter B, Dennis M, et al.
PLoS Genetics
A Selective Sweep on a Deleterious Mutation in CPT1A in Arctic Populations (2014)
Clemente FJ, Cardona A, Inchley C, Peter BM et al.
AJHG
Surfing waves of data in San Diego: sophisticated analyses provide a broad view of human genetic diversity. (2014)
Reppell M, Koch E, Peter BM, and Novembre J
Genome Biology
Detecting Range Expansions from Genetic Data (2013)
Peter BM and Slatkin M
Evolution
Distinguishing selection from standing variation from selection on a de novo mutation (2012)
Peter BM, Huerta-Sánchez E, and Nielsen, R
PLoS Genetics
Distinguishing between population bottleneck and population subdivision by a Bayesian model choice procedure. (2010)
Peter BM, Wegmann D, and Excoffier L
Molecular Ecology